zarr_url = "https://objects.eodc.eu/e05ab01a9d56408d82ac32d69a5aae2a:sample-data/tutorial_data/cpm_v253/S2B_MSIL1C_20250113T103309_N0511_R108_T32TLQ_20250113T122458.zarr"Accessing EOPF Sentinel Zarr using Rarr
Introduction
This notebook demonstrates how to retrieve remotely stored Zarr data using the Rarr package in R. We will explore how to read and visualize Zarr data (zarrays) and their metadata. For subsequent analysis and visualization we turn the zarray into stars objects which are more suitable for spatial data.
This notebook has a sibling, which demonstrates how to access the same data using the GDAL Zarr driver. You can find it here.
What we will learn
- ✏️ How to edit URLs of Zarr stores to make them readable for GDAL
- 🔎 Which read-functions and arguments to use in
starsandterra - 🚧 Current limitations of these packages
Prerequisites
To follow this notebook, you will need to load a STAC item in Zarr format from the EOPF Zarr STAC Catalog. If you are new to STAC inside the R environment, we suggest to follow the Access the EOPF Zarr STAC API with R tutorial. The item we are using for this example mirrors the one used in the Sentinel-2 L1C MSI Zarr Product Exploration notebook which is hosted on EODC’s object storage and is accessible under the URL below.
Import packages
library(Rarr) # reading; BiocManager::install("Rarr")
library(stars) # spatial objects
library(dplyr) # table operations
library(jsonlite) # parsing metadata
library(mapview) # interactive mapsMetadata exploration with Rarr
In this notebook we show the current capabilities and limitations of Rarr in terms of reading Zarr array(s) and their metadata. Successful attempts are marked with a ✅, failed attempts with a ⛔.
Using the Rarr::zarr_overview function we can get meta data for all assets in the remote Zarr store as a table. Here list all arrays and their data type, compression, dimensions, etc.
zarr_content = zarr_overview(zarr_url, as_data_frame = T) |>
mutate(var = sub(zarr_url, "", path)) |>
select(var, everything(), -path)
filter(zarr_content, grepl("reflectance", var)) var nchunks data_type compressor dim
1 /measurements/reflectance/r10m/b02 36 uint16 blosc 10980, 10980
2 /measurements/reflectance/r10m/b03 36 uint16 blosc 10980, 10980
3 /measurements/reflectance/r10m/b04 36 uint16 blosc 10980, 10980
4 /measurements/reflectance/r10m/b08 36 uint16 blosc 10980, 10980
5 /measurements/reflectance/r10m/x 1 int64 blosc 10980
6 /measurements/reflectance/r10m/y 1 int64 blosc 10980
7 /measurements/reflectance/r20m/b05 36 uint16 blosc 5490, 5490
8 /measurements/reflectance/r20m/b06 36 uint16 blosc 5490, 5490
9 /measurements/reflectance/r20m/b07 36 uint16 blosc 5490, 5490
10 /measurements/reflectance/r20m/b11 36 uint16 blosc 5490, 5490
11 /measurements/reflectance/r20m/b12 36 uint16 blosc 5490, 5490
12 /measurements/reflectance/r20m/b8a 36 uint16 blosc 5490, 5490
13 /measurements/reflectance/r20m/x 1 int64 blosc 5490
14 /measurements/reflectance/r20m/y 1 int64 blosc 5490
15 /measurements/reflectance/r60m/b01 36 uint16 blosc 1830, 1830
16 /measurements/reflectance/r60m/b09 36 uint16 blosc 1830, 1830
17 /measurements/reflectance/r60m/b10 36 uint16 blosc 1830, 1830
18 /measurements/reflectance/r60m/x 1 int64 blosc 1830
19 /measurements/reflectance/r60m/y 1 int64 blosc 1830
chunk_dim
1 1830, 1830
2 1830, 1830
3 1830, 1830
4 1830, 1830
5 10980
6 10980
7 915, 915
8 915, 915
9 915, 915
10 915, 915
11 915, 915
12 915, 915
13 5490
14 5490
15 305, 305
16 305, 305
17 305, 305
18 1830
19 1830We can see that all Sentinel-2 bands are present, but split into directories (or groups) based on spatial resolution (10, 20, 60 meters), which have different dimensions (number and size of pixels).
var nchunks data_type
1 /conditions/geometry/angle 1 unicode224
2 /conditions/geometry/band 1 unicode96
3 /conditions/geometry/detector 1 int64
4 /conditions/geometry/mean_sun_angles 1 float64
5 /conditions/geometry/mean_viewing_incidence_angles 1 float64
6 /conditions/geometry/sun_angles 1 float64
7 /conditions/geometry/viewing_incidence_angles 4 float64
8 /conditions/geometry/x 1 int64
9 /conditions/geometry/y 1 int64
10 /conditions/mask/detector_footprint/r10m/b02 36 uint8
11 /conditions/mask/detector_footprint/r10m/b03 36 uint8
12 /conditions/mask/detector_footprint/r10m/b04 36 uint8
13 /conditions/mask/detector_footprint/r10m/b08 36 uint8
14 /conditions/mask/detector_footprint/r10m/x 1 int64
15 /conditions/mask/detector_footprint/r10m/y 1 int64
16 /conditions/mask/detector_footprint/r20m/b05 36 uint8
17 /conditions/mask/detector_footprint/r20m/b06 36 uint8
18 /conditions/mask/detector_footprint/r20m/b07 36 uint8
19 /conditions/mask/detector_footprint/r20m/b11 36 uint8
20 /conditions/mask/detector_footprint/r20m/b12 36 uint8
21 /conditions/mask/detector_footprint/r20m/b8a 36 uint8
22 /conditions/mask/detector_footprint/r20m/x 1 int64
23 /conditions/mask/detector_footprint/r20m/y 1 int64
24 /conditions/mask/detector_footprint/r60m/b01 36 uint8
25 /conditions/mask/detector_footprint/r60m/b09 36 uint8
26 /conditions/mask/detector_footprint/r60m/b10 36 uint8
27 /conditions/mask/detector_footprint/r60m/x 1 int64
28 /conditions/mask/detector_footprint/r60m/y 1 int64
29 /conditions/mask/l1c_classification/r60m/b00 36 uint8
30 /conditions/mask/l1c_classification/r60m/x 1 int64
31 /conditions/mask/l1c_classification/r60m/y 1 int64
32 /conditions/meteorology/cams/aod1240 1 float32
33 /conditions/meteorology/cams/aod469 1 float32
34 /conditions/meteorology/cams/aod550 1 float32
35 /conditions/meteorology/cams/aod670 1 float32
36 /conditions/meteorology/cams/aod865 1 float32
37 /conditions/meteorology/cams/bcaod550 1 float32
38 /conditions/meteorology/cams/duaod550 1 float32
39 /conditions/meteorology/cams/isobaricInhPa 1 float64
40 /conditions/meteorology/cams/latitude 1 float64
41 /conditions/meteorology/cams/longitude 1 float64
42 /conditions/meteorology/cams/number 1 int64
43 /conditions/meteorology/cams/omaod550 1 float32
44 /conditions/meteorology/cams/ssaod550 1 float32
45 /conditions/meteorology/cams/step 1 int64
46 /conditions/meteorology/cams/suaod550 1 float32
47 /conditions/meteorology/cams/surface 1 float64
48 /conditions/meteorology/cams/time 1 int64
49 /conditions/meteorology/cams/valid_time 1 int64
50 /conditions/meteorology/cams/z 1 float32
51 /conditions/meteorology/ecmwf/isobaricInhPa 1 float64
52 /conditions/meteorology/ecmwf/latitude 1 float64
53 /conditions/meteorology/ecmwf/longitude 1 float64
54 /conditions/meteorology/ecmwf/msl 1 float32
55 /conditions/meteorology/ecmwf/number 1 int64
56 /conditions/meteorology/ecmwf/r 1 float32
57 /conditions/meteorology/ecmwf/step 1 int64
58 /conditions/meteorology/ecmwf/surface 1 float64
59 /conditions/meteorology/ecmwf/tco3 1 float32
60 /conditions/meteorology/ecmwf/tcwv 1 float32
61 /conditions/meteorology/ecmwf/time 1 int64
62 /conditions/meteorology/ecmwf/u10 1 float32
63 /conditions/meteorology/ecmwf/v10 1 float32
64 /conditions/meteorology/ecmwf/valid_time 1 int64
65 /measurements/reflectance/r10m/b02 36 uint16
66 /measurements/reflectance/r10m/b03 36 uint16
67 /measurements/reflectance/r10m/b04 36 uint16
68 /measurements/reflectance/r10m/b08 36 uint16
69 /measurements/reflectance/r10m/x 1 int64
70 /measurements/reflectance/r10m/y 1 int64
71 /measurements/reflectance/r20m/b05 36 uint16
72 /measurements/reflectance/r20m/b06 36 uint16
73 /measurements/reflectance/r20m/b07 36 uint16
74 /measurements/reflectance/r20m/b11 36 uint16
75 /measurements/reflectance/r20m/b12 36 uint16
76 /measurements/reflectance/r20m/b8a 36 uint16
77 /measurements/reflectance/r20m/x 1 int64
78 /measurements/reflectance/r20m/y 1 int64
79 /measurements/reflectance/r60m/b01 36 uint16
80 /measurements/reflectance/r60m/b09 36 uint16
81 /measurements/reflectance/r60m/b10 36 uint16
82 /measurements/reflectance/r60m/x 1 int64
83 /measurements/reflectance/r60m/y 1 int64
84 /quality/l1c_quicklook/r10m/band 1 int64
85 /quality/l1c_quicklook/r10m/tci 108 uint8
86 /quality/l1c_quicklook/r10m/x 1 int64
87 /quality/l1c_quicklook/r10m/y 1 int64
88 /quality/mask/r10m/b02 36 uint8
89 /quality/mask/r10m/b03 36 uint8
90 /quality/mask/r10m/b04 36 uint8
91 /quality/mask/r10m/b08 36 uint8
92 /quality/mask/r10m/x 1 int64
93 /quality/mask/r10m/y 1 int64
94 /quality/mask/r20m/b05 36 uint8
95 /quality/mask/r20m/b06 36 uint8
96 /quality/mask/r20m/b07 36 uint8
97 /quality/mask/r20m/b11 36 uint8
98 /quality/mask/r20m/b12 36 uint8
99 /quality/mask/r20m/b8a 36 uint8
100 /quality/mask/r20m/x 1 int64
101 /quality/mask/r20m/y 1 int64
102 /quality/mask/r60m/b01 36 uint8
103 /quality/mask/r60m/b09 36 uint8
104 /quality/mask/r60m/b10 36 uint8
105 /quality/mask/r60m/x 1 int64
106 /quality/mask/r60m/y 1 int64
compressor dim chunk_dim
1 blosc 2 2
2 blosc 13 13
3 blosc 7 7
4 blosc 2 2
5 blosc 13, 2 13, 2
6 blosc 2, 23, 23 2, 23, 23
7 blosc 13, 7, 2.... 7, 4, 2,....
8 blosc 23 23
9 blosc 23 23
10 blosc 10980, 10980 1830, 1830
11 blosc 10980, 10980 1830, 1830
12 blosc 10980, 10980 1830, 1830
13 blosc 10980, 10980 1830, 1830
14 blosc 10980 10980
15 blosc 10980 10980
16 blosc 5490, 5490 915, 915
17 blosc 5490, 5490 915, 915
18 blosc 5490, 5490 915, 915
19 blosc 5490, 5490 915, 915
20 blosc 5490, 5490 915, 915
21 blosc 5490, 5490 915, 915
22 blosc 5490 5490
23 blosc 5490 5490
24 blosc 1830, 1830 305, 305
25 blosc 1830, 1830 305, 305
26 blosc 1830, 1830 305, 305
27 blosc 1830 1830
28 blosc 1830 1830
29 blosc 1830, 1830 305, 305
30 blosc 1830 1830
31 blosc 1830 1830
32 blosc 9, 9 9, 9
33 blosc 9, 9 9, 9
34 blosc 9, 9 9, 9
35 blosc 9, 9 9, 9
36 blosc 9, 9 9, 9
37 blosc 9, 9 9, 9
38 blosc 9, 9 9, 9
39 <NA>
40 blosc 9 9
41 blosc 9 9
42 <NA>
43 blosc 9, 9 9, 9
44 blosc 9, 9 9, 9
45 <NA>
46 blosc 9, 9 9, 9
47 <NA>
48 <NA>
49 <NA>
50 blosc 9, 9 9, 9
51 <NA>
52 blosc 9 9
53 blosc 9 9
54 blosc 9, 9 9, 9
55 <NA>
56 blosc 9, 9 9, 9
57 <NA>
58 <NA>
59 blosc 9, 9 9, 9
60 blosc 9, 9 9, 9
61 <NA>
62 blosc 9, 9 9, 9
63 blosc 9, 9 9, 9
64 <NA>
65 blosc 10980, 10980 1830, 1830
66 blosc 10980, 10980 1830, 1830
67 blosc 10980, 10980 1830, 1830
68 blosc 10980, 10980 1830, 1830
69 blosc 10980 10980
70 blosc 10980 10980
71 blosc 5490, 5490 915, 915
72 blosc 5490, 5490 915, 915
73 blosc 5490, 5490 915, 915
74 blosc 5490, 5490 915, 915
75 blosc 5490, 5490 915, 915
76 blosc 5490, 5490 915, 915
77 blosc 5490 5490
78 blosc 5490 5490
79 blosc 1830, 1830 305, 305
80 blosc 1830, 1830 305, 305
81 blosc 1830, 1830 305, 305
82 blosc 1830 1830
83 blosc 1830 1830
84 blosc 3 3
85 blosc 3, 10980.... 1, 1830,....
86 blosc 10980 10980
87 blosc 10980 10980
88 blosc 10980, 10980 1830, 1830
89 blosc 10980, 10980 1830, 1830
90 blosc 10980, 10980 1830, 1830
91 blosc 10980, 10980 1830, 1830
92 blosc 10980 10980
93 blosc 10980 10980
94 blosc 5490, 5490 915, 915
95 blosc 5490, 5490 915, 915
96 blosc 5490, 5490 915, 915
97 blosc 5490, 5490 915, 915
98 blosc 5490, 5490 915, 915
99 blosc 5490, 5490 915, 915
100 blosc 5490 5490
101 blosc 5490 5490
102 blosc 1830, 1830 305, 305
103 blosc 1830, 1830 305, 305
104 blosc 1830, 1830 305, 305
105 blosc 1830 1830
106 blosc 1830 1830Now that we have explored the content, let’s get some related metadata. Rarr provides the read_zattrs function for this purpose, which looks for the .zattrs file inside the Zarr store.
⛔ Unfortunately we were not successful when applying it to our remote store.
read_zattrs(zarr_url)Error in read_zattrs(zarr_url): The group or array does not contain attributes (.zattrs)read_zattrs(file.path(zarr_url, ".zattrs"))Error in read_zattrs(file.path(zarr_url, ".zattrs")): The group or array does not contain attributes (.zattrs)✅ As a simple workaround we can read the JSON directly:
zattrs = read_json(file.path(zarr_url, ".zattrs"))
names(zattrs)[1] "other_metadata" "other_metadata4" "stac_discovery" var nchunks data_type
1 /conditions/geometry/angle 1 unicode224
2 /conditions/geometry/band 1 unicode96
3 /conditions/geometry/detector 1 int64
4 /conditions/geometry/mean_sun_angles 1 float64
5 /conditions/geometry/mean_viewing_incidence_angles 1 float64
6 /conditions/geometry/sun_angles 1 float64
7 /conditions/geometry/viewing_incidence_angles 4 float64
8 /conditions/geometry/x 1 int64
9 /conditions/geometry/y 1 int64
10 /conditions/mask/detector_footprint/r10m/b02 36 uint8
11 /conditions/mask/detector_footprint/r10m/b03 36 uint8
12 /conditions/mask/detector_footprint/r10m/b04 36 uint8
13 /conditions/mask/detector_footprint/r10m/b08 36 uint8
14 /conditions/mask/detector_footprint/r10m/x 1 int64
15 /conditions/mask/detector_footprint/r10m/y 1 int64
16 /conditions/mask/detector_footprint/r20m/b05 36 uint8
17 /conditions/mask/detector_footprint/r20m/b06 36 uint8
18 /conditions/mask/detector_footprint/r20m/b07 36 uint8
19 /conditions/mask/detector_footprint/r20m/b11 36 uint8
20 /conditions/mask/detector_footprint/r20m/b12 36 uint8
21 /conditions/mask/detector_footprint/r20m/b8a 36 uint8
22 /conditions/mask/detector_footprint/r20m/x 1 int64
23 /conditions/mask/detector_footprint/r20m/y 1 int64
24 /conditions/mask/detector_footprint/r60m/b01 36 uint8
25 /conditions/mask/detector_footprint/r60m/b09 36 uint8
26 /conditions/mask/detector_footprint/r60m/b10 36 uint8
27 /conditions/mask/detector_footprint/r60m/x 1 int64
28 /conditions/mask/detector_footprint/r60m/y 1 int64
29 /conditions/mask/l1c_classification/r60m/b00 36 uint8
30 /conditions/mask/l1c_classification/r60m/x 1 int64
31 /conditions/mask/l1c_classification/r60m/y 1 int64
32 /conditions/meteorology/cams/aod1240 1 float32
33 /conditions/meteorology/cams/aod469 1 float32
34 /conditions/meteorology/cams/aod550 1 float32
35 /conditions/meteorology/cams/aod670 1 float32
36 /conditions/meteorology/cams/aod865 1 float32
37 /conditions/meteorology/cams/bcaod550 1 float32
38 /conditions/meteorology/cams/duaod550 1 float32
39 /conditions/meteorology/cams/isobaricInhPa 1 float64
40 /conditions/meteorology/cams/latitude 1 float64
41 /conditions/meteorology/cams/longitude 1 float64
42 /conditions/meteorology/cams/number 1 int64
43 /conditions/meteorology/cams/omaod550 1 float32
44 /conditions/meteorology/cams/ssaod550 1 float32
45 /conditions/meteorology/cams/step 1 int64
46 /conditions/meteorology/cams/suaod550 1 float32
47 /conditions/meteorology/cams/surface 1 float64
48 /conditions/meteorology/cams/time 1 int64
49 /conditions/meteorology/cams/valid_time 1 int64
50 /conditions/meteorology/cams/z 1 float32
51 /conditions/meteorology/ecmwf/isobaricInhPa 1 float64
52 /conditions/meteorology/ecmwf/latitude 1 float64
53 /conditions/meteorology/ecmwf/longitude 1 float64
54 /conditions/meteorology/ecmwf/msl 1 float32
55 /conditions/meteorology/ecmwf/number 1 int64
56 /conditions/meteorology/ecmwf/r 1 float32
57 /conditions/meteorology/ecmwf/step 1 int64
58 /conditions/meteorology/ecmwf/surface 1 float64
59 /conditions/meteorology/ecmwf/tco3 1 float32
60 /conditions/meteorology/ecmwf/tcwv 1 float32
61 /conditions/meteorology/ecmwf/time 1 int64
62 /conditions/meteorology/ecmwf/u10 1 float32
63 /conditions/meteorology/ecmwf/v10 1 float32
64 /conditions/meteorology/ecmwf/valid_time 1 int64
65 /measurements/reflectance/r10m/b02 36 uint16
66 /measurements/reflectance/r10m/b03 36 uint16
67 /measurements/reflectance/r10m/b04 36 uint16
68 /measurements/reflectance/r10m/b08 36 uint16
69 /measurements/reflectance/r10m/x 1 int64
70 /measurements/reflectance/r10m/y 1 int64
71 /measurements/reflectance/r20m/b05 36 uint16
72 /measurements/reflectance/r20m/b06 36 uint16
73 /measurements/reflectance/r20m/b07 36 uint16
74 /measurements/reflectance/r20m/b11 36 uint16
75 /measurements/reflectance/r20m/b12 36 uint16
76 /measurements/reflectance/r20m/b8a 36 uint16
77 /measurements/reflectance/r20m/x 1 int64
78 /measurements/reflectance/r20m/y 1 int64
79 /measurements/reflectance/r60m/b01 36 uint16
80 /measurements/reflectance/r60m/b09 36 uint16
81 /measurements/reflectance/r60m/b10 36 uint16
82 /measurements/reflectance/r60m/x 1 int64
83 /measurements/reflectance/r60m/y 1 int64
84 /quality/l1c_quicklook/r10m/band 1 int64
85 /quality/l1c_quicklook/r10m/tci 108 uint8
86 /quality/l1c_quicklook/r10m/x 1 int64
87 /quality/l1c_quicklook/r10m/y 1 int64
88 /quality/mask/r10m/b02 36 uint8
89 /quality/mask/r10m/b03 36 uint8
90 /quality/mask/r10m/b04 36 uint8
91 /quality/mask/r10m/b08 36 uint8
92 /quality/mask/r10m/x 1 int64
93 /quality/mask/r10m/y 1 int64
94 /quality/mask/r20m/b05 36 uint8
95 /quality/mask/r20m/b06 36 uint8
96 /quality/mask/r20m/b07 36 uint8
97 /quality/mask/r20m/b11 36 uint8
98 /quality/mask/r20m/b12 36 uint8
99 /quality/mask/r20m/b8a 36 uint8
100 /quality/mask/r20m/x 1 int64
101 /quality/mask/r20m/y 1 int64
102 /quality/mask/r60m/b01 36 uint8
103 /quality/mask/r60m/b09 36 uint8
104 /quality/mask/r60m/b10 36 uint8
105 /quality/mask/r60m/x 1 int64
106 /quality/mask/r60m/y 1 int64
compressor dim chunk_dim
1 blosc 2 2
2 blosc 13 13
3 blosc 7 7
4 blosc 2 2
5 blosc 13, 2 13, 2
6 blosc 2, 23, 23 2, 23, 23
7 blosc 13, 7, 2.... 7, 4, 2,....
8 blosc 23 23
9 blosc 23 23
10 blosc 10980, 10980 1830, 1830
11 blosc 10980, 10980 1830, 1830
12 blosc 10980, 10980 1830, 1830
13 blosc 10980, 10980 1830, 1830
14 blosc 10980 10980
15 blosc 10980 10980
16 blosc 5490, 5490 915, 915
17 blosc 5490, 5490 915, 915
18 blosc 5490, 5490 915, 915
19 blosc 5490, 5490 915, 915
20 blosc 5490, 5490 915, 915
21 blosc 5490, 5490 915, 915
22 blosc 5490 5490
23 blosc 5490 5490
24 blosc 1830, 1830 305, 305
25 blosc 1830, 1830 305, 305
26 blosc 1830, 1830 305, 305
27 blosc 1830 1830
28 blosc 1830 1830
29 blosc 1830, 1830 305, 305
30 blosc 1830 1830
31 blosc 1830 1830
32 blosc 9, 9 9, 9
33 blosc 9, 9 9, 9
34 blosc 9, 9 9, 9
35 blosc 9, 9 9, 9
36 blosc 9, 9 9, 9
37 blosc 9, 9 9, 9
38 blosc 9, 9 9, 9
39 <NA>
40 blosc 9 9
41 blosc 9 9
42 <NA>
43 blosc 9, 9 9, 9
44 blosc 9, 9 9, 9
45 <NA>
46 blosc 9, 9 9, 9
47 <NA>
48 <NA>
49 <NA>
50 blosc 9, 9 9, 9
51 <NA>
52 blosc 9 9
53 blosc 9 9
54 blosc 9, 9 9, 9
55 <NA>
56 blosc 9, 9 9, 9
57 <NA>
58 <NA>
59 blosc 9, 9 9, 9
60 blosc 9, 9 9, 9
61 <NA>
62 blosc 9, 9 9, 9
63 blosc 9, 9 9, 9
64 <NA>
65 blosc 10980, 10980 1830, 1830
66 blosc 10980, 10980 1830, 1830
67 blosc 10980, 10980 1830, 1830
68 blosc 10980, 10980 1830, 1830
69 blosc 10980 10980
70 blosc 10980 10980
71 blosc 5490, 5490 915, 915
72 blosc 5490, 5490 915, 915
73 blosc 5490, 5490 915, 915
74 blosc 5490, 5490 915, 915
75 blosc 5490, 5490 915, 915
76 blosc 5490, 5490 915, 915
77 blosc 5490 5490
78 blosc 5490 5490
79 blosc 1830, 1830 305, 305
80 blosc 1830, 1830 305, 305
81 blosc 1830, 1830 305, 305
82 blosc 1830 1830
83 blosc 1830 1830
84 blosc 3 3
85 blosc 3, 10980.... 1, 1830,....
86 blosc 10980 10980
87 blosc 10980 10980
88 blosc 10980, 10980 1830, 1830
89 blosc 10980, 10980 1830, 1830
90 blosc 10980, 10980 1830, 1830
91 blosc 10980, 10980 1830, 1830
92 blosc 10980 10980
93 blosc 10980 10980
94 blosc 5490, 5490 915, 915
95 blosc 5490, 5490 915, 915
96 blosc 5490, 5490 915, 915
97 blosc 5490, 5490 915, 915
98 blosc 5490, 5490 915, 915
99 blosc 5490, 5490 915, 915
100 blosc 5490 5490
101 blosc 5490 5490
102 blosc 1830, 1830 305, 305
103 blosc 1830, 1830 305, 305
104 blosc 1830, 1830 305, 305
105 blosc 1830 1830
106 blosc 1830 1830The same information can be retrieved with sf::gdal_utils('mdiminfo', ...).
Reading Zarr data with Rarr
Reading zarrays from a remote Zarr store with Rarr and turning them into useful spatial R objects is not as straightforward as with GDAL-based stars and terra functions. We will demonstrate the current capabilities and limitations of Rarr by using a reference object retrieved through the GDAL Zarr driver which is loaded in the background.
# GDAL-based reference object
zz_gdalstars object with 2 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
b01 0.1299 0.1542 0.1628 0.3541124 0.4304 1.6556
dimension(s):
from to offset delta refsys x/y
x 1 1830 3e+05 60 WGS 84 / UTM zone 32N [x]
y 1 1830 5e+06 -60 WGS 84 / UTM zone 32N [y]We start with an naive attempt to read the entire Zarr store using Rarr::read_zarr_array.
⛔ Rarr, like stars and terra, is not capable of reading multiple arrays from the remote Zarr store simultaneously.
read_zarr_array(zarr_url)Error: ✅ But if a single band array is selected we are able to access the desired information.
band_variable = "/measurements/reflectance/r60m/b01"
zarray = read_zarr_array(paste0(zarr_url, band_variable))
str(zarray) int [1:1830, 1:1830] 9548 8117 7158 5906 7007 7523 5999 6570 7165 6666 ...Raster geometry
The function returns a matrix (2D array) lacking any spatial metadata (e.g. Coordinate Reference System) or map coordinates. Let’s plot it together with the reference object for comparison.
image(zarray)
image(zz_gdal$b01)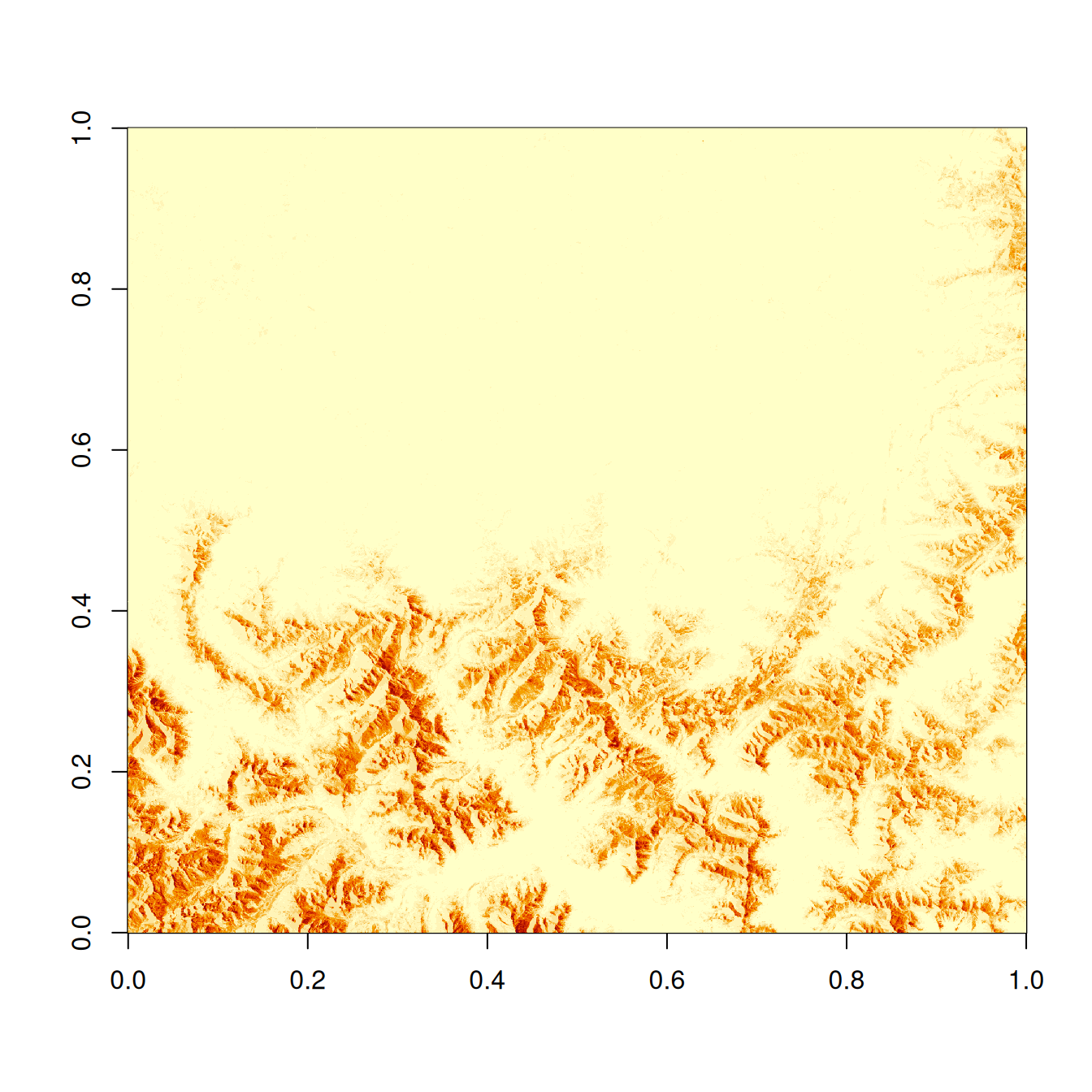

The raw zarray has flipped X- and Y-dimensions! But we can access X and Y coordinates for each pixel separately and handle the issue by transposing them later (using t()).
# retrieve X and Y coordinates
zarray_60m_x = paste0(zarr_url, "/measurements/reflectance/r60m/x") |>
read_zarr_array()
zarray_60m_y = paste0(zarr_url, "/measurements/reflectance/r60m/y") |>
read_zarr_array()
print(range(zarray_60m_x))[1] 300030 409770print(range(zarray_60m_y))[1] 4890270 5000010Regardless of their ordering, are the coordinate values consistent with those retrieved via GDAL?
print(range(st_get_dimension_values(zz_gdal, "x")))[1] 300030 409770print(range(st_get_dimension_values(zz_gdal, "y")))[1] 4890270 5000010✅ Yes!
Reflectance values
Before building the spatial object we should also check if the actual data values are consistent between both methods.
head(t(zarray)[1,]) # flipped zarray[1] 9548 8117 7158 5906 7007 7523head(zz_gdal$b01[1,]) # reference arrray[1] 0.8548 0.7117 0.6158 0.4906 0.6007 0.6523By comapring the first few values of each array we can observe 2 important differences:
zarraycontains integers values whilezz_gdalhas scaled values (float) with factor 10000.- after rescaling, min and max for example still differ (0.1299 vs. 0.2299 / 1.6556 vs. 1.7556), indicating an offset of 0.1.
💡 Explanation: The data provider applies scale and offset to store more memory efficient integer values. A scale factor of 10000 preserves 4 decimals, while an offset of 0.1 avoids division by 0. When reading raw data like that we need to apply scale and offset manually.
This infrormation is important metadata, but currently not accessible via Rarr. At this point we can only find it by digging deep into the metadata retrieved by GDAL:
# GDAL-comaptible VSI URL
vsi_url = paste0("ZARR:/vsicurl/", dplyr::as_label(zarr_url))
# get entire metadata
info = sf::gdal_utils("mdiminfo", source = vsi_url, quiet = T) |> fromJSON()
# access scale and offset for 60 meter reflectance bands
(scale = info$groups$measurements$groups$reflectance$groups$r60m$arrays$b01$scale)[1] 1e-04(offset = info$groups$measurements$groups$reflectance$groups$r60m$arrays$b01$offset)[1] -0.1Constructing a stars object from zarray components
Let’s combine all pieces of the puzzle to form a spatial object: - data array - scale and offset - X and Y coordinates - CRS
# read CRS from Zarr attributes
zarr_crs = zattrs$stac_discovery$properties$`proj:epsg`
# band name
var = basename(band_variable)
# read and transpose data matrix
# apply scale and offset
# convert to stars
zz = st_as_stars(t(zarray) * scale + offset) |>
st_set_dimensions(1, names = "x", values = zarray_60m_x, is_raster = TRUE) |>
st_set_dimensions(2, names = "y", values = zarray_60m_y, is_raster = TRUE) |>
setNames(var)
# assign CRS
st_crs(zz) = st_crs(zarr_crs)
# print the object
zzstars object with 2 dimensions and 1 attribute
attribute(s), summary of first 1e+05 cells:
Min. 1st Qu. Median Mean 3rd Qu. Max.
b01 0.1299 0.1542 0.1628 0.3541124 0.4304 1.6556
dimension(s):
from to offset delta refsys point x/y
x 1 1830 3e+05 60 WGS 84 / UTM zone 32N FALSE [x]
y 1 1830 5e+06 -60 WGS 84 / UTM zone 32N FALSE [y]We can compare the newly constructed object with reference read via GDAL. They should have identical geometry and values.
identical(zz[[1]] |> str(),
zz_gdal[[1]] |> str()) num [1:1830, 1:1830] 0.855 0.753 0.7 0.756 0.718 ...
num [1:1830, 1:1830] 0.855 0.753 0.7 0.756 0.718 ...[1] TRUELooks good! Let’s visualize both objects.
# plot as maps
plot(zz, axes = TRUE)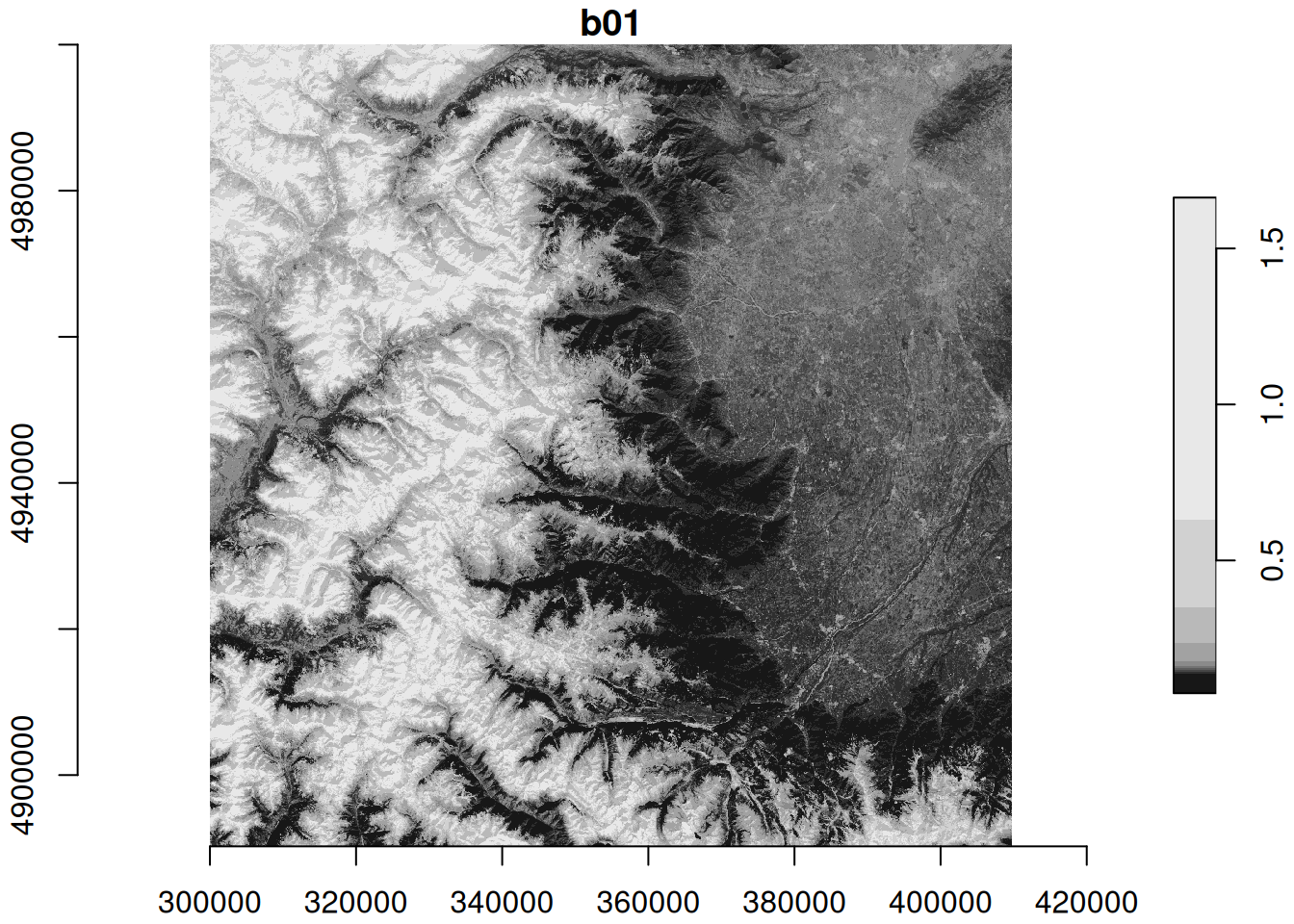
plot(zz_gdal, axes = TRUE)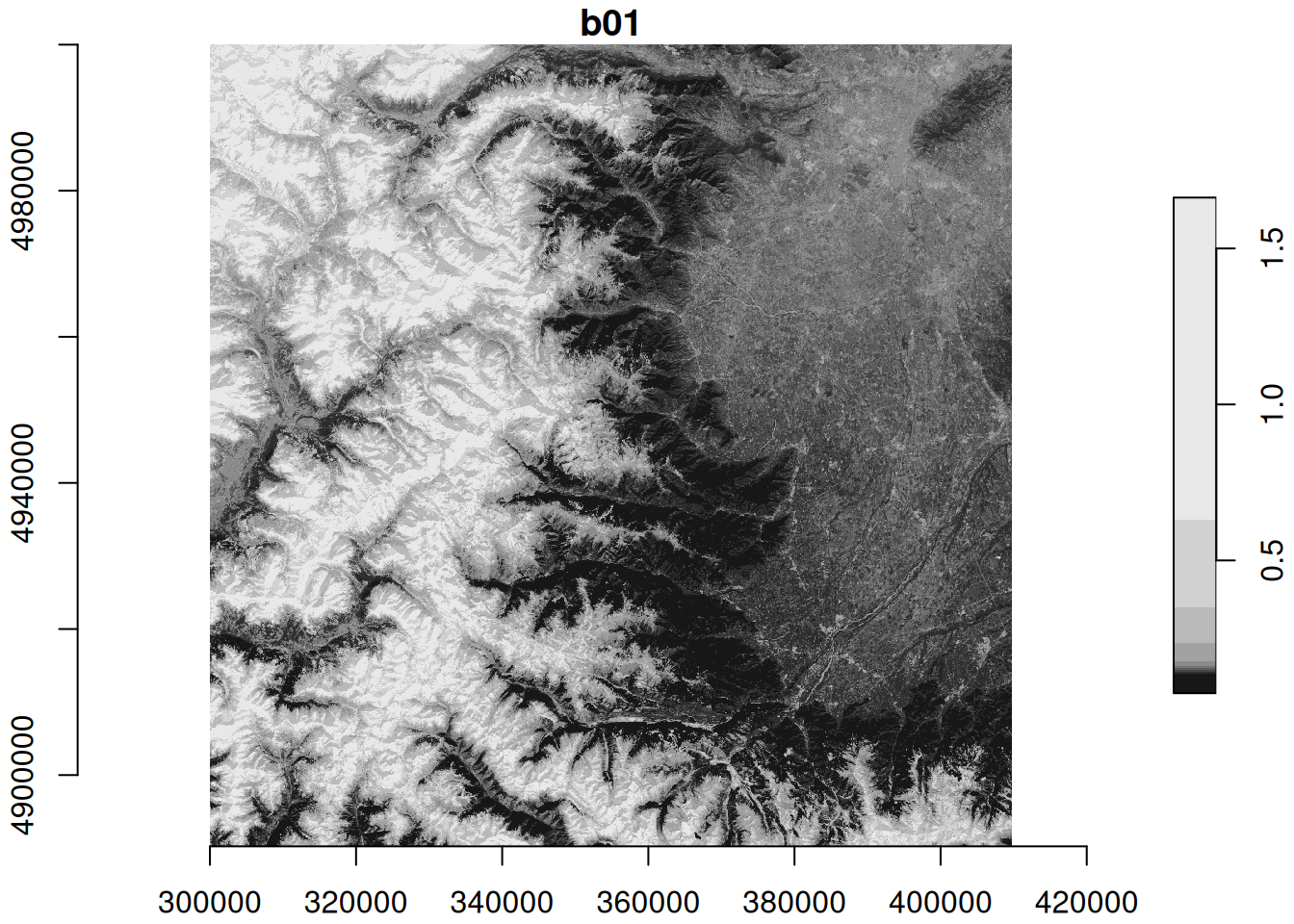
The identical maps show Sentinel-2 band 1 (coastal aerosol band) reflectance at 60m spatial resolution for north-western Italy with the Alps in the West and the city of Torino in the North-East.
Wrapping it all up in a function
We can enclose these steps in a single function with some flexibility for variable names and resolutions. This allows us to retrieve any band at any available resolution from the remote Zarr store. Since scale and offset are not accessible via Rarr, we need to provide them as function arguments.
st_read_zarray = function(path, var, res, scale = 1, offset = 0, ...){
# room for checks and input tests:
# stopifnot valid zarr url / variable / resolution ...
# get metadata including CRS
zattrs = jsonlite::read_json(file.path(path, ".zattrs"))
zarr_crs = zattrs$stac_discovery$properties$`proj:epsg`
# zattrs$stac_discovery$properties$`eopf:resolutions`
res_char = switch(as.character(res),
"10" = "r10m",
"20" = "r20m",
"60" = "r60m",
stop("Resolution must be one of 10, 20, or 60.")
)
# ...: make use of index arguments of read_zarr_array if desired
zarray = file.path(path, "measurements/reflectance", res_char, var) |>
read_zarr_array(...)
zarray_x = file.path(path, "measurements/reflectance", res_char, "x") |>
read_zarr_array(...)
zarray_y = file.path(path, "measurements/reflectance", res_char, "y") |>
read_zarr_array(...)
# transpose matrix before converting to stars
z = st_as_stars(t(zarray) * scale + offset) |>
st_set_dimensions(1, names = "x", values = zarray_60m_x, is_raster = TRUE) |>
st_set_dimensions(2, names = "y", values = zarray_60m_y, is_raster = TRUE) |>
setNames(var)
st_crs(z) = st_crs(zarr_crs)
return(z)
}Read and combine multiple zarrays
✅ Now we can read multiple bands, combine them to a single multi-band object, and visualize them in a static map…
system.time({
b01 = st_read_zarray(zarr_url, "b01", 60, scale=0.0001, offset = 0.1)
b09 = st_read_zarray(zarr_url, "b09", 60, scale=0.0001, offset = 0.1)
multi_band = c(b01, b09) |> merge(name="band") |> setNames("reflectance")
}) user system elapsed
4.042 0.066 20.762 plot(multi_band, axes = TRUE)downsample set to 1
… or by generating an interactive visualization:
mapview(multi_band)Benchmark
Using Rarr to read remote Zarr stores is just one of several options. R packages stars and terra provide functionality, that uses GDAl’s Zarr driver in the background. To see if there is a benefit to each of the methods, we can compare their execution time and memory allocation with a little benchmark.
# GDAL-compatible VSI URL
vsi_prefix = "ZARR:/vsicurl/"
vsi_url = paste0(vsi_prefix, dplyr::as_label(zarr_url))
# functions for reading a single Sentinel-2 band to memory
f_stars = function(){rs = read_stars(paste(vsi_url, band_variable, sep = ":"))}
f_mdim = function(){sm = read_mdim(vsi_url, band_variable, proxy = FALSE) |>
setNames(basename(band_variable))}
f_rast = function(){tr = terra::rast(vsi_url, band_variable) |> terra::toMemory()}
f_rarr = function(){rr = st_read_zarray(zarr_url, "b01", 60)}bench::mark(f_stars(), f_mdim(), f_rast(), f_rarr(),
check = F, iterations = 5)# A tibble: 4 × 6
expression min median `itr/sec` mem_alloc `gc/sec`
<bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
1 f_stars() 8.2s 8.38s 0.120 25.6MB 0
2 f_mdim() 7.64s 7.79s 0.124 26.3MB 0.0247
3 f_rast() 8.26s 8.47s 0.119 83.2KB 0
4 f_rarr() 11.1s 11.54s 0.0869 148.5MB 0.0869The benchmark does not reveal a clear winner in terms of speed. However, Rarr seems not to be faster than GDAL but allocates more memory compared to stars and terra.
💪 Now it is your turn
- 🔭 Task: Supply more arguments to
st_read_zarrayand pass a subset to read only parts on an array. Visit?Rarr::read_zarr_arrayfor more details. - 💾 Task: Try writing zarrays to disk using
Rarr::write_zarr_array(). - 📖 Read the Rarr documentation.
Conclusion
In this notebook we have demonstrated how to access remote Zarr stores in R using the Rarr package. We have seen that it is possible to read specific data arrays, but read_zarr_array lacks the ability of accessing anything other than the raw array. To combine it with a CRS or coordinates we need to leverage a spatial class like stars and manually set the relevant metadata. Further, the benchmark did not reveal a significant benefit regarding processing time or memory allocation over “traditional” GDAL-based data retrieval.
What’s next?
Further tests should evaluate wether data retrieved via Rarr versus GDAL are identical in terms of spatial extent and offset.
Explore this related notebook which demonstrates how to access the same Zarr data using the GDAL Zarr driver via the stars and terra packages.